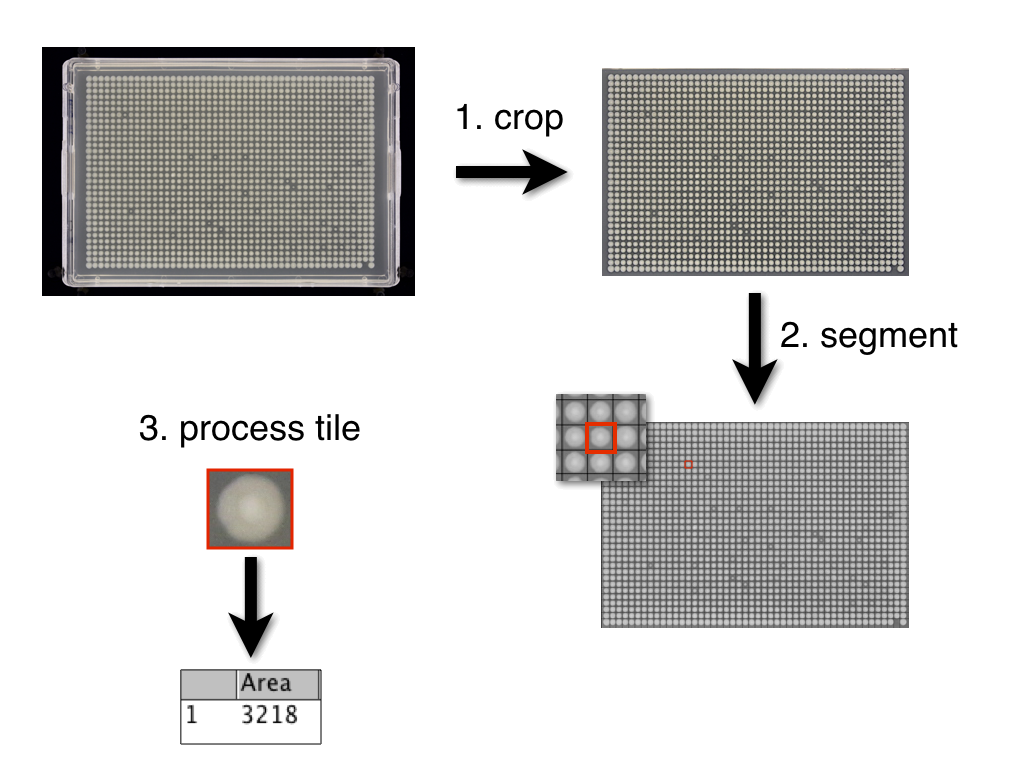
Image analysis workflow
Generally, Iris takes the following steps to analyze colonies:
- crops image (to keep only colony array)
- cuts (segments) cropped image into tiny images (tiles), each holding one colony
- analyzes each tile independently to get size other phenotypic characteristics
Adjusting image analysis workflow parameters
This is done by editing a provided user settings file in JSON format. Iris needs to be re-run for any changes in the file to take effect.
Global settings
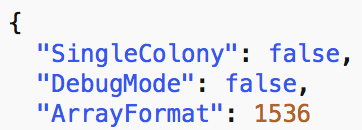
ArrayFormat
Colony array density.
Valid formats are:
24, 96, 384, 1536, 6144
SingleColony mode is intended to be used when providing a folder with single-colony pictures. This mode will bypass cropping and segmentation steps, and feed the pictures directly on the TileReaders defined by the selected profile for tile processing (step #3 on schematic above).
If SingleColony mode is set, ArrayFormat is ignored.
DebugMode will provide a verbose output.
Profile settings
The JSON file provided in the distribution has examples for some profiles. To add profile-settings for other/new profiles, one can simply copy the settings from another profile (within the brackets) and edit the profile name.
Upon loading, Iris will report the profiles for which settings are defined in the user settings file.

ProfileName the name of the profile to be adjusted.
rotationSettings defines whether the image will be automatically detected for the colony array to be perfectly horizontal. Users may choose to bypass the automatic detection for a defined rotation (in degrees).
croppingSettings defines if the image will be automatically cropped to keep just the colonies. Alternatively a fixed cropping area will be applied, based on the defined coordinates:
X, Y-start define the top left corner of the picture that includes the colonies.
X, Y-end define the width and height of the final cropped picture.
segmentationSettings defines whether colony tile boundaries will be adjusted to include the entire colony (and the maximum adjustment distance in pixels).
detectionSettings defines the minimum colony size and circularity thresholds under which the detected object will not be considered a colony.
Iris design
Iris was designed in a modular fashion; in other words, the above worflow steps are separate modules that one can plug in, modify, and replace at will.
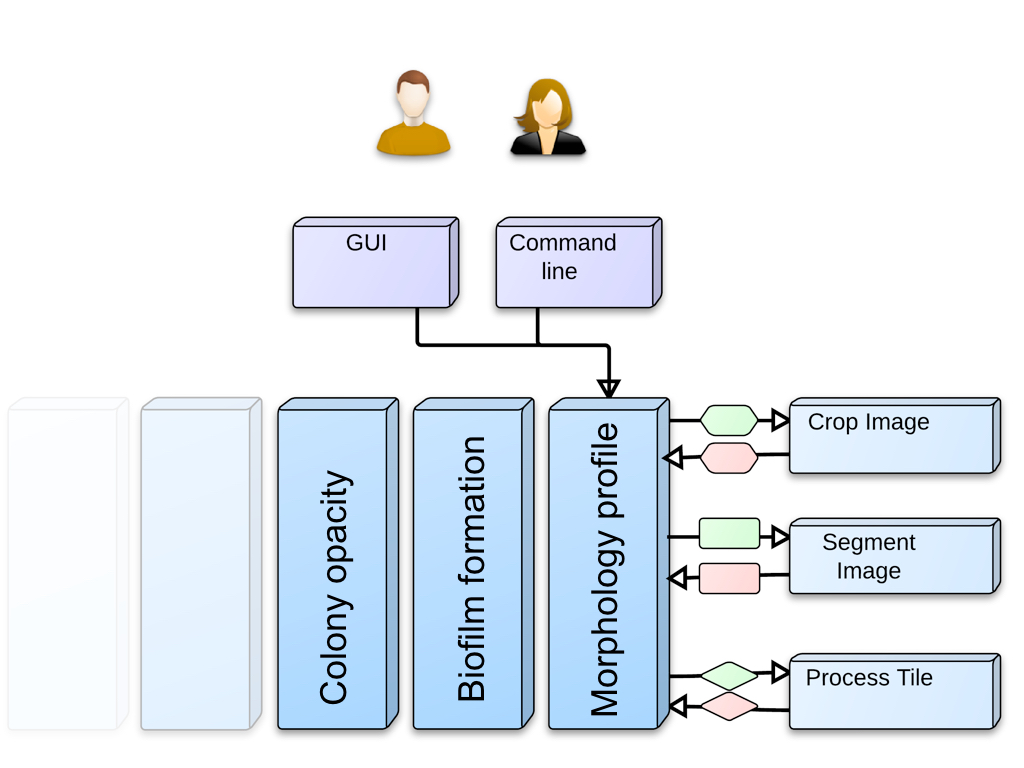
module interoperability
To help new/modified modules connect in the same way to the profile class, I use connecting classes, such as TileReaderInput and TileReaderOutput (illustrated as green and pink shapes in the above)
These connecting classes only need to be extended if you add new readouts not already covered by existing connecting subclasses.
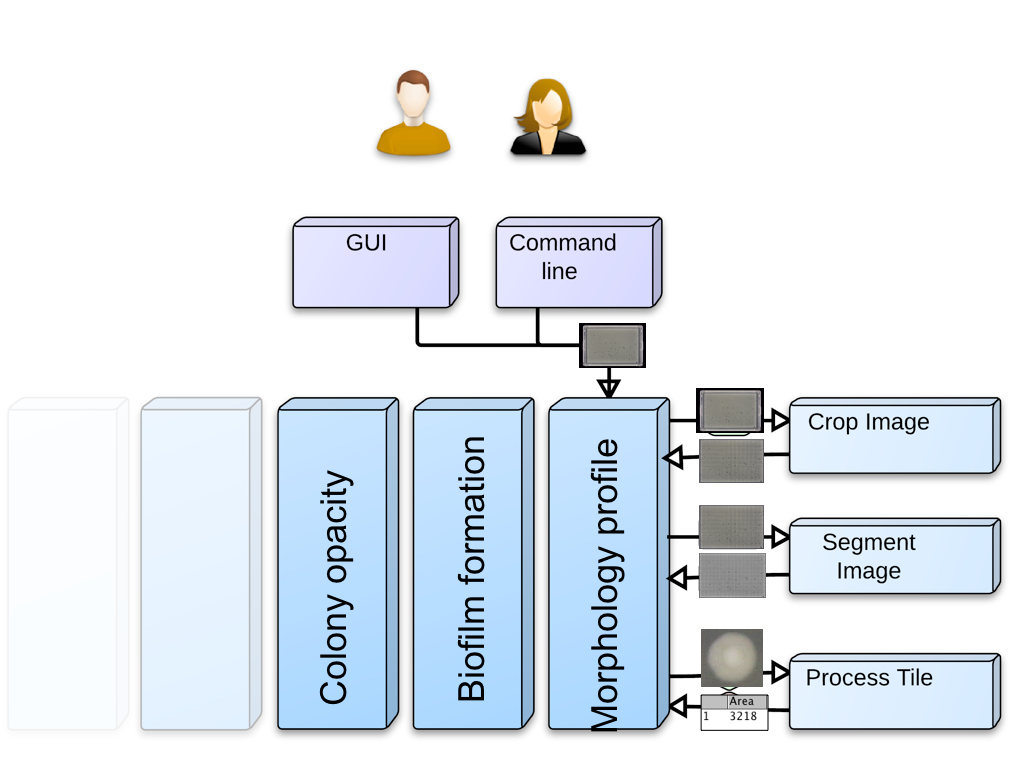
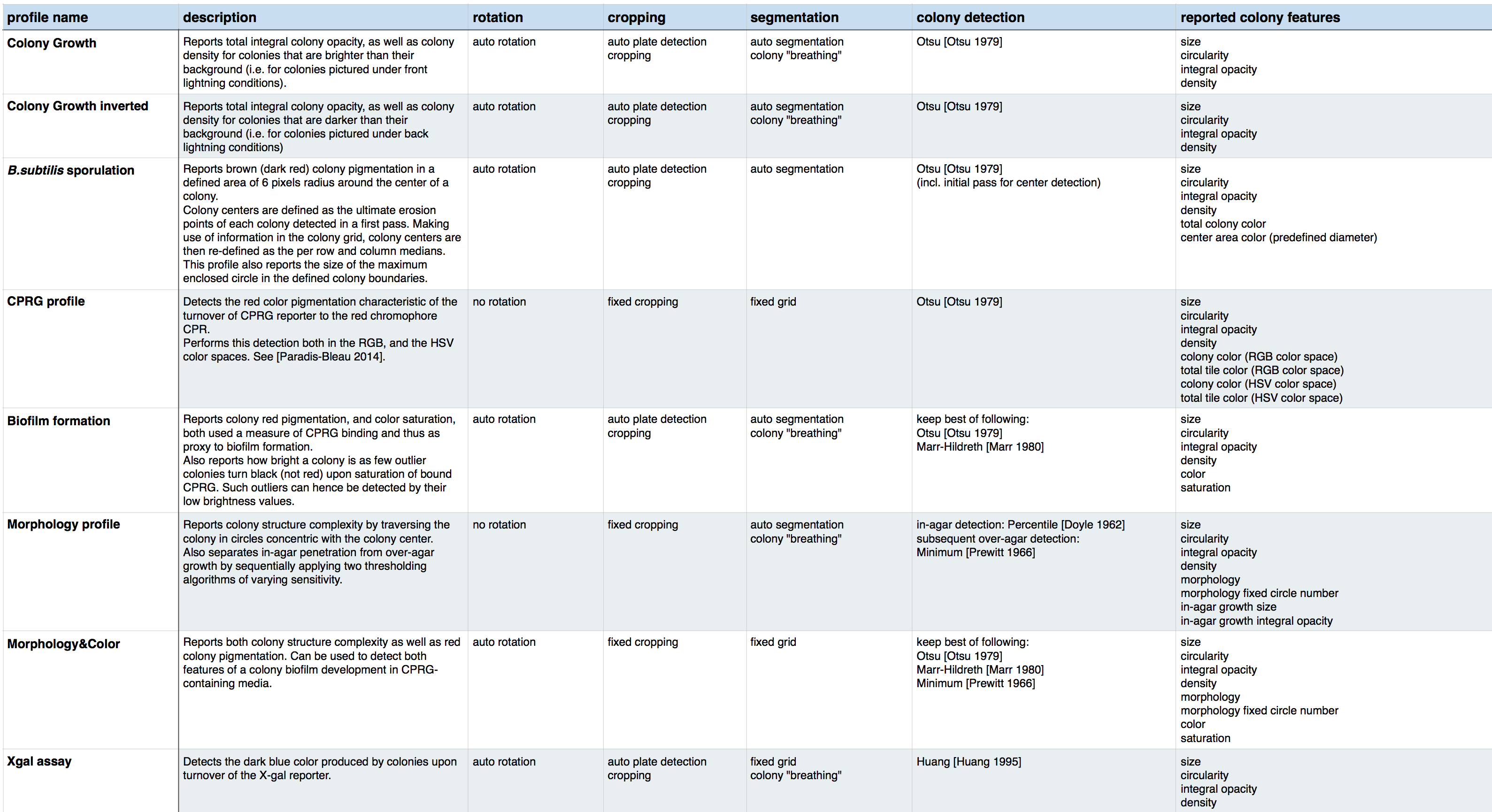
Algorithms used per profile
Each profile calls different modules (as illustrated above), but different profiles can call on the same modules.
Here is a table listing the algorithms used per profile:
I have questions regarding Iris development
Please post a question on the Iris Q&A forum